Module simulator#
How the simulator works?#
Simulator is a class that allows to perform previously designed experiment. To run it we need a network (multilayer or temporal) (note that it can as well have one layer) and corresponding model. After the experiment is completed, user is able to see results in form of report and visualisation of global states of the nodes.
Example of usage#
Initialise multilayer network from nx predefined network:
import networkx as nx from network_diffusion.mln.mlnetwork import MultilayerNetwork network = MultilayerNetwork() names = ['illness', 'awareness', 'vaccination'] network.from_nx_layer(nx.les_miserables_graph(), names)
Initialise propagation model and set possible transitions with probabilities:
model = PropagationModel() phenomenas = [('S', 'I', 'R'), ('UA', 'A'), ('UV', 'V')] for l, p in zip(names, phenomenas): model.add(l, p) model.compile(background_weight=0.005) model.set_transition('illness.S', 'illness.I', ['vaccination.UV', 'awareness.UA'], 0.9) model.set_transition('illness.S', 'illness.I', ['vaccination.V', 'awareness.A'], 0.05) model.set_transition('illness.S', 'illness.I', ['vaccination.UV', 'awareness.A'], 0.2) model.set_transition('illness.I', 'illness.R', ['vaccination.UV', 'awareness.UA'], 0.1) model.set_transition('illness.I', 'illness.R', ['vaccination.V', 'awareness.A'], 0.7) model.set_transition('illness.I', 'illness.R', ['vaccination.UV', 'awareness.A'], 0.3) model.set_transition('vaccination.UV', 'vaccination.V', ['awareness.A', 'illness.S'], 0.03) model.set_transition('vaccination.UV', 'vaccination.V', ['awareness.A', 'illness.I'], 0.01) model.set_transition('awareness.UA', 'awareness.A', ['vaccination.UV', 'illness.S'], 0.05) model.set_transition('awareness.UA', 'awareness.A', ['vaccination.V', 'illness.S'], 1) model.set_transition('awareness.UA', 'awareness.A', ['vaccination.UV', 'illness.I'], 0.2) model.set_transition('illness', (('awareness.UA', 'illness.R', 'vaccination.UV'), ('awareness.UA', 'illness.I', 'vaccination.UV')), 0.7)
3. Initialise initial parameters of propagation in network. Parameters’ names must correspond with names in model and network. Numbers in tuples describe how many nodes has which local state (in alphabetic order):
phenomenas = {'illness': (70, 6, 1), 'awareness': (60, 17), 'vaccination': (70, 7)}
4. Perform propagation experiment. Propagation lasts as many epochs as defined (here 200). After the experiment, Logger object is returned where logs are being stored:
experiment = Simulator(model, network)
experiment.set_initial_states(phenomenas)
logs = experiment.perform_propagation(200)
5. Save experiment results. User is able to save them to file or print out to the console:
logs.report(to_file=True, path=getcwd()+'/results', visualisation=True)
Logs contain:
- description of the network (txt file)
- description of the propagation model (txt file)
- propagation report in all phenomena (separate csv file for each)
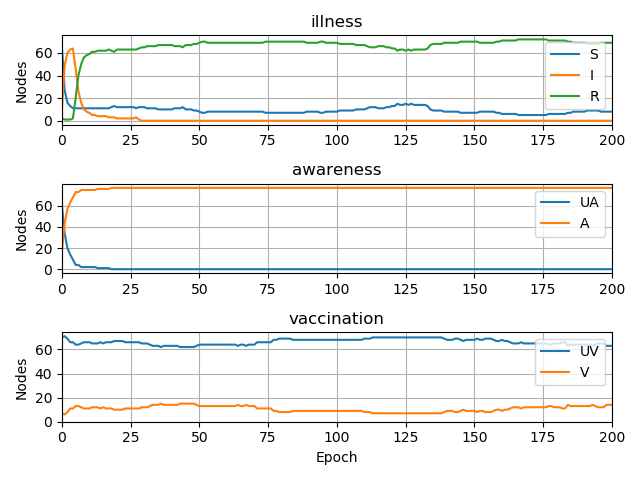
- visualisation of propagation